Superresolution with Allele's photoswitchable tags
mMaple and its predecessor mClavGR2 are monomeric green-to-red photoswitchable fluorescent proteins optimized for use in fusion protein localization tracking and superresolution microscopy using the PALM technique. mMaple also tends to display substantially better performace as a fusion tag than other photoconvertible proteins such as mEos2, reducing the risk of localization artifacts. mMaple may also be used for SIM imaging.
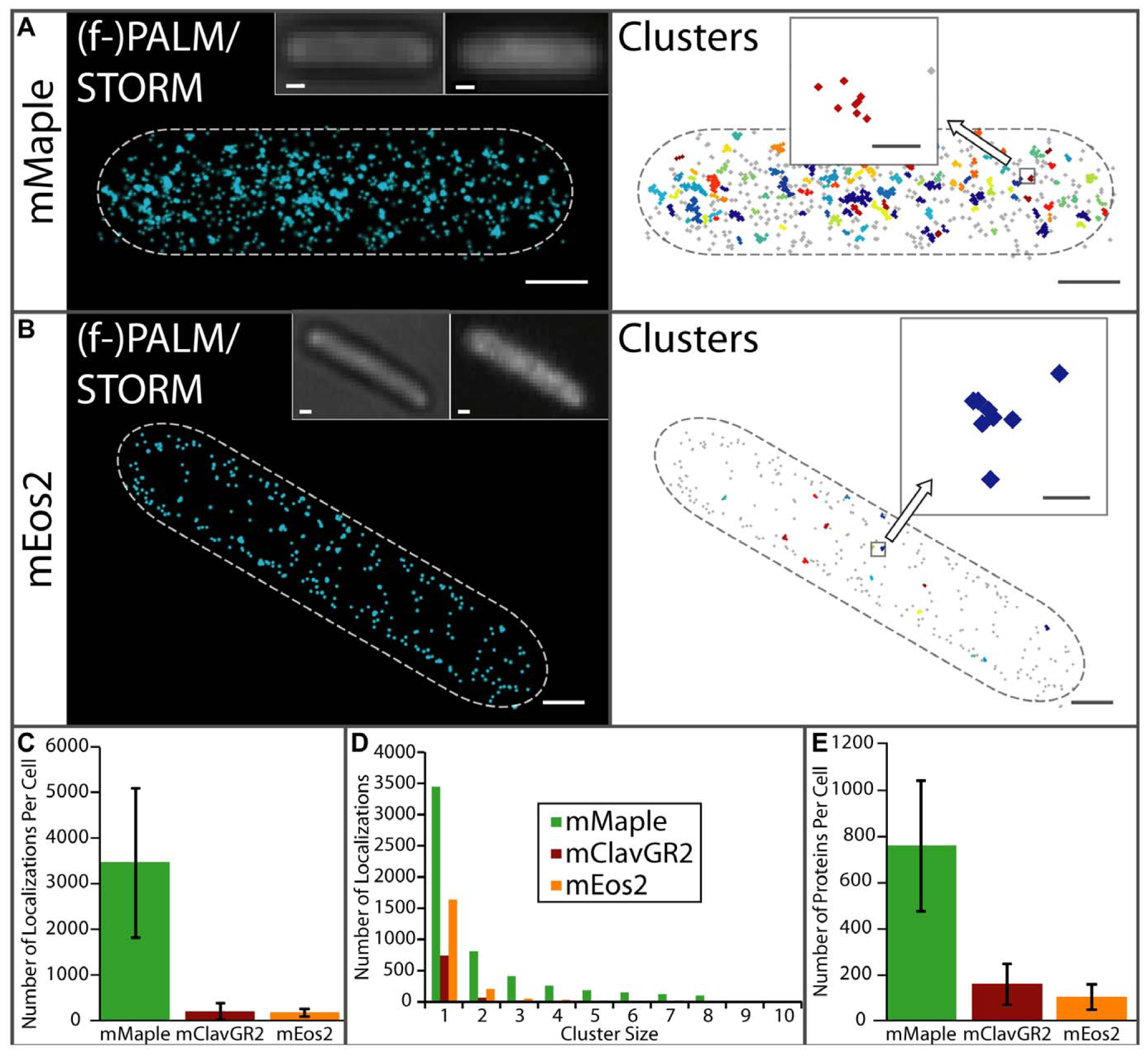
Below is an example of PALM imaging of individual cytoplasmic mMaple molecules in E. coli, illustrating improved performance over mEos2. Localizations are represented as normalized 2D Gaussian peaks with widths given by their theoretical localization precisions (left panels) and plotted as small markers grouped into clusters with adjacent spacing of 30 nm or less (right panels). Individual protein localizations are shown in grey whereas closely spaced localizations (less than 30 nm) are grouped into clusters of the same color (right panels). Conventional bright field and widefield fluorescence images are inset. Scale bars are 500 nm. Statistics for number of proteins localized and clusters are shown in the lower panels. Full details on superresolution imaging using mMaple can be found in the original publication (Download PDF).